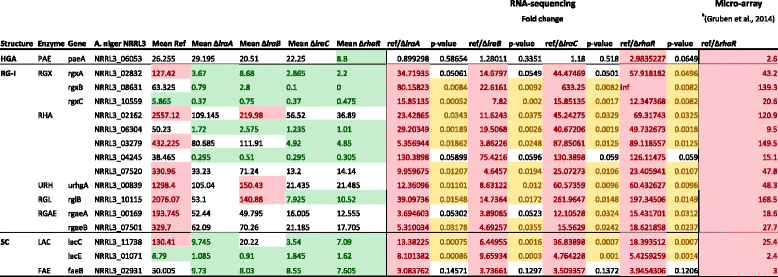
- Based on Gruben, B.S., M. Zhou, A. Wiebenga, J. Ballering, K.M. Overkamp, P.J. Punt & R.P. de Vries, (2014) Aspergillus niger RhaR, a regulator involved in L-rhamnose release and catabolism. Appl Microbiol Biotechnol 98: 5531-5540
- The expression levels are mean values of duplicate samples. Genes with values higher than 120 are considered highly expressed and marked red.
- Genes with values lower than 20 are considered low expressed and marked green.
- The fold change is the difference in expression between the reference strain and the deletion mutants.
- The cutoff for differential expression is fold change >1.5 (cells marked red if upregulated and green if downregulated) and p-value <0.05 (cells marked yellow).
- HGA homogalacturonan, RG-I Rhamnogalacturonan-I, SC side chains, PAE = pectin acetyl esterase, RGX = exorhamnogalacturonase
- RHA = endorhamnogalacturonase, URH = unsaturated rhamnogalacturonyl hydrolase, RGL = rhamnogalacturonan lyase
- RGAE = rhamnogalacturonan acetyl esterase, LAC = beta-galactosidase, FAE = feruloyl esterase.

