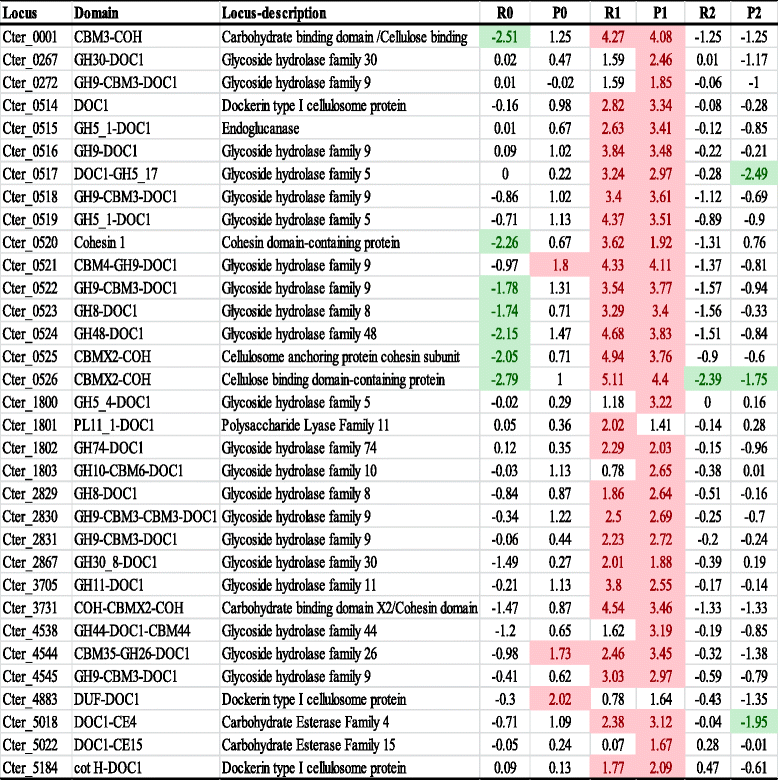
- Transcriptomic Z-scores Rnet are represented as: (i) R0: xylose grown cells – cellobiose grown cells; (ii) R1: α-cellulose grown cells – cellobiose grown cells; and (iii) R2: xylan grown cells – cellobiose grown cells. Proteomic Z-scores Pnet are represented as: (i) P0: xylose grown cells – cellobiose grown cells; (ii) P1: α-cellulose grown cells – cellobiose grown cells; and (iii) P2: xylan grown cells – cellobiose grown cells. Z-scores of ≥1.65, up regulated in the corresponding substrate with respect to cellobiose are highlighted in red. Z-scores of ≤ −1.65, down regulated in the corresponding substrate with respect to cellobiose are highlighted in green. For any protein or RNA transcripts, a negative value represents higher expression on cellobiose, while a positive value represents higher expression in any of the other corresponding substrate. For any protein or RNA transcripts, a negative value represents higher expression on cellobiose, while a positive value represents higher expression in any of the other corresponding substrate. DUF: domain of unknown function; Cot H: spore coat protein H; DOC1: dockerin type 1; COH: cohesion domain

